Brief description
The experimental activities carried out at the BioChem Lab in Rome are focused on the study of the effects of legacy and emerging organic micropollutants on organisms belonging to different levels of the trophic web (from microbial communities to higher organisms) in the context of ongoing climate change. The analytical determinations on the biota are aimed at evaluating bioaccumulation and biomagnification processes of target pollutants. Other studies performed at the BioChem Lab focus on:
- The degradation rates (evaluation of the half-life time - DT50 - and determination of metabolites or transformation products of target compounds) of the organic contaminants by setting up laboratory-scale experiments.
- Evaluate the effects of organic micropollutants on specific target organisms (e.g. avoidance behaviour test in Eisenia foetida) and on the structure (PhosphoLipid Fatty Acid analysis-PLFA) and function (Community Level Physiological Profile - CLPP) of autochthonous microbial communities, through the preparation of laboratory-scale experiments and environmental monitoring.
- Investigate the occurrence and diffusion of antibiotic resistance genes (ARGs) and the intI1 gene which is an anthropogenic impact proxy, among natural microbial communities related to the presence of antibiotics in environmental matrices.
Environmental compartments
The bioaccumulation/biomagnification studies of organic micropollutants involve biota such as fish, molluscs, terrestrial and aquatic earthworms and terrestrial and aquatic plant species. The studies on persistence and effects of pollutants on natural microbial communities are related to different environmental compartments such as surface waters and sediments (seas, rivers, lakes), snow/ice and soils.
Analytical techniques
Studies on bioaccumulation/biomagnification/persistence of organic contaminants are performed by a combination of:
- Pre-treatment methods (e.g. freeze-drying, filtration, etc.).
- Extraction/Clean-up methods (solid-phase extraction-SPE, pressurized liquid extraction-PLE, liquid-liquid extraction-LL).
- Sensitive and selective analytical methods based on the coupling of chromatographic techniques (HPLC or GC) and fluorescence, FID-ECD, and mass spectrometric (MS) detection.
- The avoidance behaviour test is based on the exposure of organisms, specifically earthworms belonging to the Eisenia foetida species, to different sub-lethal concentrations of organic contaminants, to evaluate their escape/avoidance behaviour in the contaminated area.
- The evaluation of the composition of natural microbial communities is carried out by PLFA (PhosphoLipid Fatty Acid) profiling. This biochemical technique includes the extraction of fatty acids from microbial membrane phospholipids in the environmental matrix and a methylation reaction followed by separation and analysis with gas chromatography.
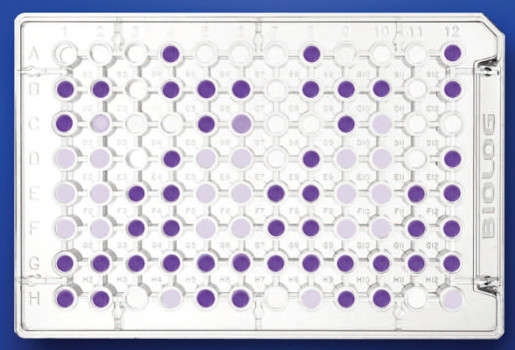
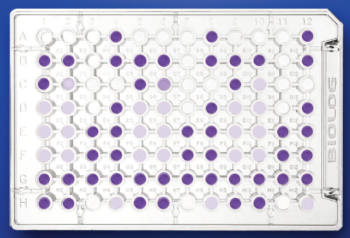
- The analysis of microbial diversity at a functional level is performed by the determination of the metabolic/physiological profile (CLPP). This physiological assay involves the incubation of the environmental sample in specific plates and spectrophotometric measurements at fixed times.
- The analysis of ARGs and intI1 gene is performed by DNA extraction from the environmental samples by using specific kits and subsequent analysis by qPCR.
Equipment
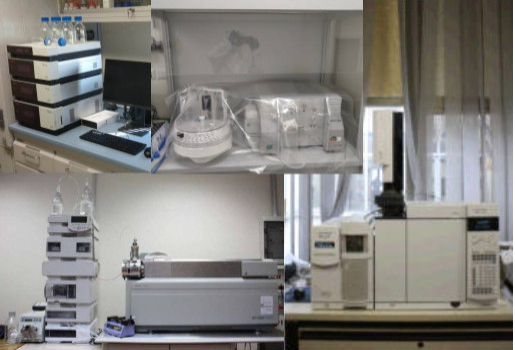
 The BioChem Lab is equipped with the following analytical tools:
The BioChem Lab is equipped with the following analytical tools:
Benchtop lyophilizer (freeze-dryer LABCONCO) 2.5 L capacity, equipped with a touchscreen display, for the pre-treatment of solid matrices subsequently extracted with PLE.
Solid Phase Extraction (SPE):12 inlets of the solid phase extractor are connected to cartridges packed with specific adsorbents for the extraction of target compounds from liquid matrices through a vacuum system.
Syncore® Analyst (Buchi) for the simultaneous pre-concentration of liquid phase samples.
Rotavapor R 100 (Buchi), equipped with an electronic interface to control the vacuum system and the recirculating chiller.
Gas Chromatograph (Perkin Elmer, Clarus 480) coupled to a FID-ECD detector (Flame Ionization Detector- Electron Capture Detector). The instrument is connected to an autosampler (Autosystem, Perkin Elmer) and is controlled by TotalChrom Software.
Gas chromatograph (Thermo Fisher, Trace 3000) coupled to a mass spectrometry (MS) detector (Thermo Fisher, ISQ7000). The device is connected to an autosampler (Thermo Fisher. AI 1310) and is controlled by a Chromeleon software.
HPLC (quaternary pump, column Oven mod. LC-100 and Micro Pump Series 200, Perkin Elmer, USA) coupled to a fluorescence detector (Perkin Elmer Series 200a). The device is controlled by Chromeleon Software.
HPLC (binary pump, Vanquish TM Core HPLC system, Thermo Scientific TM, Italy) coupled to a high-resolution mass spectrometer (Orbitrap Exploris 120, Thermo Scientific TM, Italy). The device is controlled by XcaliburSoftware (version 5.1).
Spectrophotometer (Biolog Microstation System, Biolog Inc.) for the reading of microplates (96 positions). This instrument is controlled by specific software.
Contact person: Dr. Luisa Patrolecco – luisa.patrolecco AT cnr.it